FOOD IN CRYPTO ARENA
Protein-protein interactions are central to understanding the functional relationship between proteins. We have curated the available information and augmented with context-specific analysis to ease your research journey.
PALWORLD CRYPTO
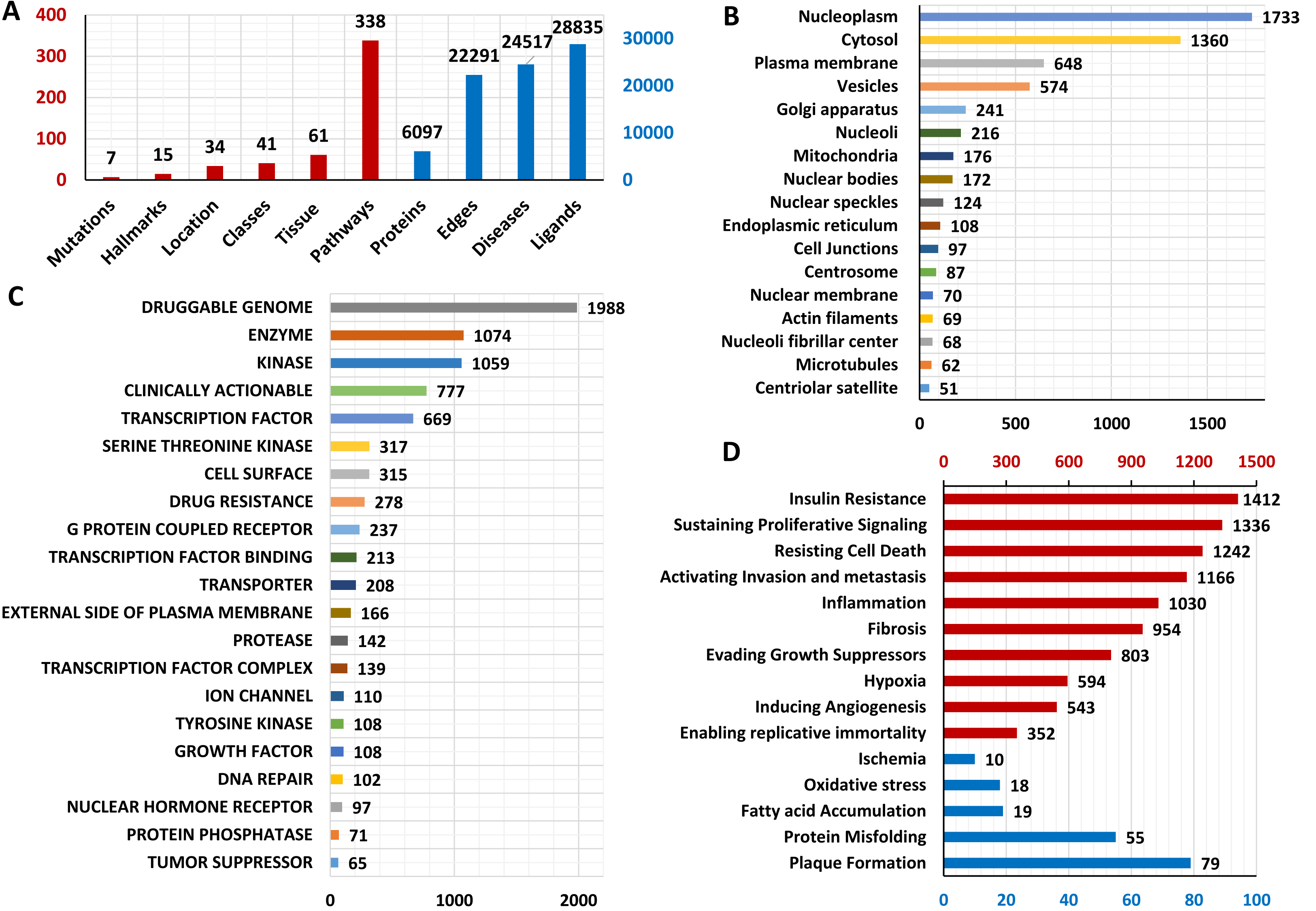
This is an indigenously developed web-based application designed by Dr. Samrat Chatterjee and his group, which can generate context-specific directional PPI networks from the input proteins and detect their biological and topological importance in the network. konnect2prot (k2p) contains data curated from various databases with multidimensional information to build a network relevant to the end user. This platform is also equipped with network algorithms that can further enhance the decision making process.
About Us
PRICE OF BLOCKCHAIN

BEST CRYPTO NEWS WEBSITES
We study biological systems in their entirety or in fragments through the lens of system biology. The former covers topics such as network biology, machine learning-based drug discovery, and so forth, while the latter covers various differential equation based kinetic models. The group is supervised by Dr. Samrat Chatterjee and consists of post doctoral fellows, PhD student and research fellows. They are mainly from mathematical and computational backgrounds keen to work on biological problems in an interdisciplinary environment.
Read MoreSome significant recent publications from the group.
FUD IN CRYPTOBLAZE CRYPTOHOW TO BUY FLOKI INU ON COINBASEBINANCE PYTHON APICRYPTO DAY TRADING SIMULATOR
CRYPTO MACHINES INGAS STATIONGEORGE CRYPTO R USCRYPTO BASE SCANNERFINK CRYPTOCROB CRYPTO
CRYPTO FUNK STRAIN LEAFLYCRYPTO TRADERWHAT DOES BURN CRYPTO MEANCRYPTO WHALE TRACKERCRYPTO BASTION 25MSHENBLOOMBERG
Features
BLUZELLE CRYPTO
Directionality
کوین مارکت گپ
Context-specificity
CAGA CRYPTO
Spreaders
COINS TO BUY RIGHT NOW
Network topology
COINBASE PRIVATE KEY
Structural information
BITBOY CRYPTO LAMBORGHINI
Enrichment analysis
LADY OF CRYPTO
Disease interactome
NO DEPOSIT CRYPTO BONUS
PTM
CRYPTO SALARY
Protein classes
BEST LONG TERM CRYPTO
Team
CRYPTO NEWS NEO
Other contributers: Ekant Sharma, Komal Sharma, Krishna
Acknowledgement
FIRST VIP WALLET

Department of Biotechnology
We would like to thank DBT for proving us the finanical support for this project.
Funding body by Govt. of India

OmniPath
We would like to thank OmniPath for providing directional relationship of protein pairs.
Directional database for PPI

Signor 2.0
We would like to thank signor for providing directional relationship of protein pairs.
Directional database for PPI

DisGeNET
We would like to thank disgenet for providing disease information related to protein.
Disease database

Kyoto Encyclopedia of Genes and Genomes (KEGG)
We would like to thank KEGG for providing information of biological pathways associated with proteins.
Biological pathway database

Gene ontology (GO)
We would like to thank GO for providing information of biological process and molecular functions associated with proteins.
Biological ontology database

The Human Protein Atlas (HPA)
We would like to thank HPA for providing information of tissue specific expression and subcellular associated with proteins.
Human Protein Atlas

Translational Health Science and Technology Institute
We would like to thank THSTI for providing the infrastructure to carry out the work.
THSTI

Uniprot
We would like to thank uniprot for providing detailed information of protein charaterstics including its functions and pathways.
Protein information database

Enrichr
We would like to thank Enrichr for providing enrichment score of pathway, functions and diseases.
Protein enrichment webserver

Protein data bank (PDB)
We would like to thank PDB for providing structure related information.
Protein structure database

The Drug Gene Interaction Database (DGIdb)
We would like to thank DGIdb for providing information of ligands and classes associated with proteins.
Drug target database

Signalink
We would like to thank signalink for providing information of signalling pathways associated with proteins.
Signalling pathway database

Database of Curated Mutations (DoCM)
We would like to thank DoCM for providing information of disease-causing mutations associated with proteins.
Disease mutation database
We would also like to pass our gratitude to open source software develpers of Flask, Python, Dash (Plotly), MongoDB.
FAQ
COINBASE ERRORS

CRYPTO CLUB ELON MUSK
We have made this far on our own, collectively we can accomplish much more !!!




